Madurella mycetomatis
The genus Madurella was originally based on tissue morphology (mycetoma with black grains) and the formation of sterile cultures on mycological media.
Initially two species were described, M. mycetomatis and M. grisea. However recent molecular studies have recognised five species: Madurella mycetomatis, Trematosphaeria grisea (formerly M. grisea), M. fahalii, M. pseudomycetomatis and M. tropicana (Desnos-Ollivier et al. 2006, de Hoog et al. 2004a, 2012). All species have been isolated from soil and are major causative agents of mycetoma.
RG-2 organism.
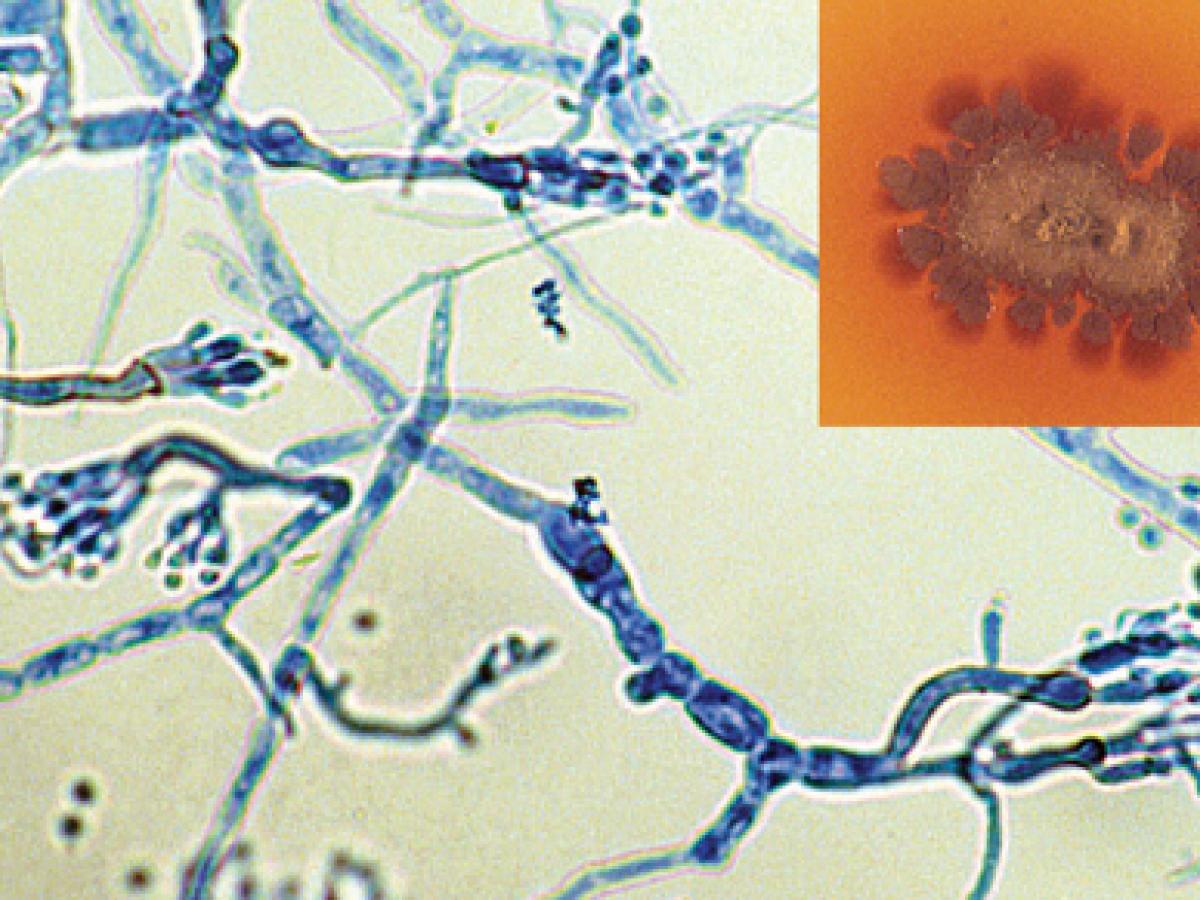
Culture showing the typical brown diffusable pigment in the agar and phialides of M. mycetomatis (rarely seen as most isolates are sterile).
Morphological description:
Colonies are slow growing, flat and leathery at first, white to yellow to yellowish-brown, becoming brownish, folded and heaped with age, and with the formation of aerial mycelia. A brown diffusible pigment is characteristically produced in primary cultures. Although most cultures are sterile, two types of conidiation have been observed, the first being flask-shaped phialides that bear rounded conidia, the second being simple or branched conidiophores bearing pyriform conidia (3-5 µm) with truncated bases. The optimum temperature for growth of this mould is 37C.
Grains of Madurella mycetomatis (tissue microcolonies) are brown or black, 0.5-1.0 mm in size, round or lobed, hard and brittle, composed of hyphae which are 2-5 µm in diameter, with terminal cells expanded to 12-15 (30) µm in diameter.
M. mycetomatis can be distinguished from Trematosphaeria grisea by growth at 37C and its inability to assimilate sucrose.
Key features:
Black grain mycetoma, growth at 37C, diffusible brown pigment produced on culture and the occasional presence of phialides.
Molecular identification:
ITS sequencing is recommended for species separation (Ahmed et al. 2014b, Desnos-Olliver et al. 2006, Irinyi et al. 2015). A five locus phylogenetic analysis was performed by Ahmed et al. (2014a) using the ITS, D1/D2, RPB2 and EF-1α genes.
References:
McGinnis (1980), Chandler et al. (1980), Rippon (1988), de Hoog et al. (2000, 2004a, 2012, 2015), Desnos-Ollivier et al. (2006).
