Candida
Recently, several taxonomic rearrangements have been made and many well-known Candida species have been renamed and moved to other genera, notably Nakaseomyces glabratus (formerly C. glabrata), Pichia kudriavzevii (formerly Candida krusei), Meyerozyma guilliermondii (formerly Candida guilliermondii), Clavispora lusitaniae (formerly Candida lusitaniae), Kluyveromyces marxianus (formerly Candida kefyr), Diutina catenulata (formerly Candida catenulata), Diutina rugosa (formerly Candida rugosa) and Wickerhamomyces anomalus (formerly Candida pelliculosa). Candida parapsilosis is also now recognised as species complex (Tavanti 2005; Correia 2006; Alcoba-Florez 2005).
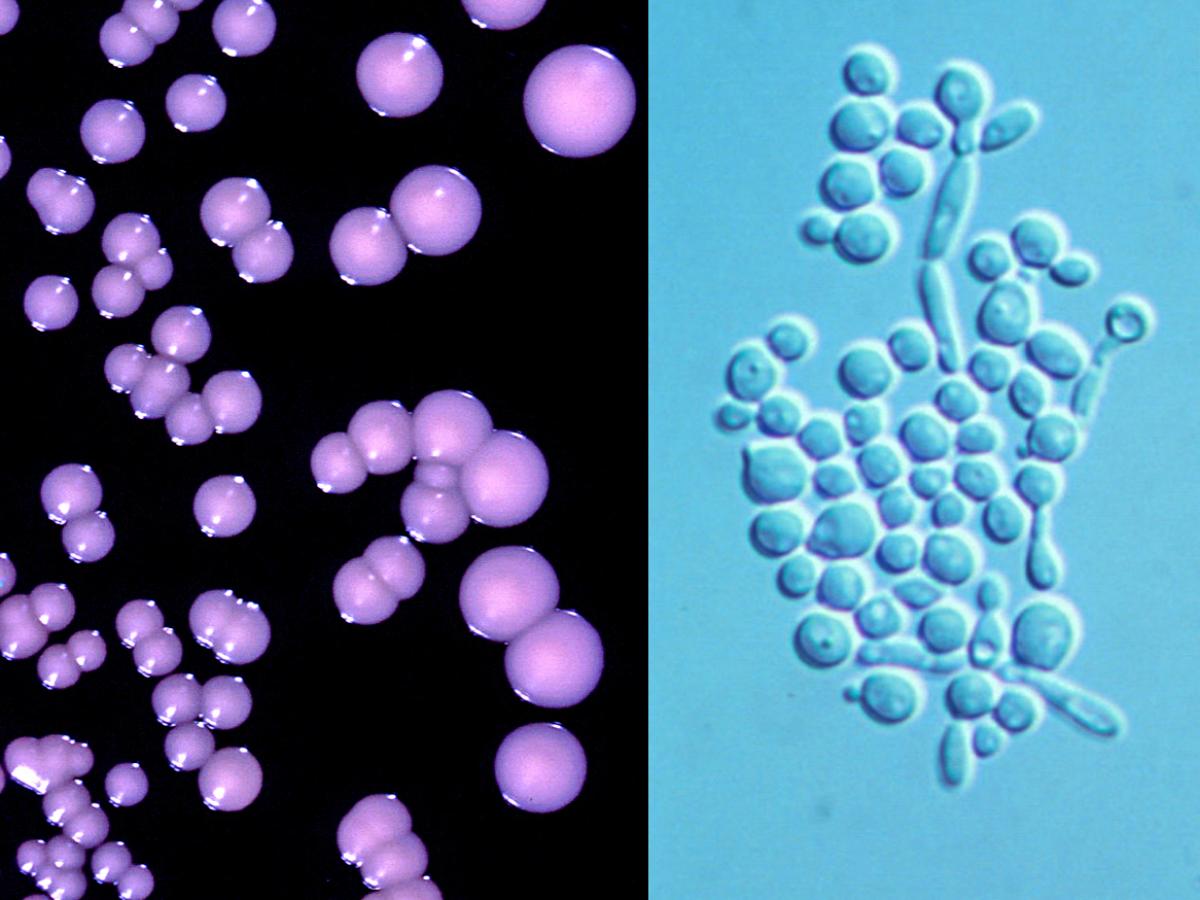
Candida albicans showing typical cream-coloured, smooth surfaced, waxy colonies and narrow based budding spherical to ovoid blastoconidia.
The genus Candida is characterised by globose to elongate yeast-like cells or blastoconidia that reproduce by narrow-based multilateral budding. Pseudohyphae and occasionally true hyphae may also be present. Colony pigmentation is usually absent. Ballistoconidia are not formed. Arthroconidia may be formed, but not extensively. Sexual reproduction is absent. Glucose may be fermented. Nitrate may be assimilated. Starch-like compounds are not produced. The diazonium blue B reaction is negative.
The genus is highly polyphyletic, as it comprises mitosporic species that are devoid of special distinguishing features (Lachance et al., 2011). Accordingly, several taxonomic rearrangements have been made and many well-known Candida species have been moved to other genera, notably Nakaseomyces glabratus (formerly C. glabrata), Pichia kudriavzevii (formerly Candida krusei), Meyerozyma guilliermondii (formerly Candida guilliermondii), Clavispora lusitaniae (formerly Candida lusitaniae), Kluyveromyces marxianus (formerly Candida kefyr), Diutina catenulata (formerly Candida catenulata), Diutina rugosa (formerly Candida rugosa) and Wickerhamomyces anomalus (formerly Candida pelliculosa). Candida parapsilosis are recognised as species complexes (Alcoba-Florez et al., 2005; Tavanti et al., 2005; Correia et al., 2006).
Several species may be aetiological agents, most commonly Candida albicans, followed by C. parapsilosis, Nakaseomyces glabratus, C. tropicalis and Pichia kudriavzevii. Altogether, these five species account for >95% of human infections. However a number of other species may also be isolated. All are ubiquitous and occur naturally on humans.
Species descriptions
-
Candida albicans
Candida albicans is a commensal of mucous membranes and the gastrointestinal tract. Environmental isolations have been made from sources contaminated by human or animal excreta, such as polluted water, soil, air and plants.
RG-2 organism.
Culture:
Colonies (SDA) white to cream-coloured smooth, glabrous, yeast-like.Microscopy:
Spherical to subspherical budding blastoconidia, 2-7 x 3-8 µm in size.India ink preparation:
Negative - no capsules present.Dalmau plate culture:
Branched pseudohyphae with dense verticils of blastoconidia. Spherical chlamydospores, mostly terminal, often on a slightly swollen subtending cell, are formed near the edge of the cover slip.Physiological Tests: + Positive, - Negative, v Variable, w Weak, s Slow, nd No Data Germ Tube + L-Sorbose v L-Arabinose v D-Glucitol v Fermentation Sucrose v D-Arabinose v 𝝰-M-D-Glucoside v Glucose + Maltose + D-Ribose v D-Gluconate v Galactose v Cellobiose - L-Rhamnose - DL-Lactate + Sucrose v Trehalose v D-Glucosamime v myo-Inositol - Maltose + Lactose - N-A-D-glucosamine v 2-K-D-Gluconate + Lactose - Melibiose - Glycerol v D-Glucuronate - Trehalose v Raffinose - Erythritol - Nitrate - Assimilation Melezitose v Ribitol v Urease - Glucose + Soluble Starch + Galactitol - 0.1% Cycloheximide + Galactose + D-Xylose + D-Mannitol + Growth at 40C + Key features:
Germ tube positive, production of chlamydospores on Dalmau plate culture, fermentation of glucose, sugar assimilation profile and a distinctive green colour on CHROMagar. Note: Germ tube negative variants (previously known as C. claussenii), and sucrose-negative variants (previously described as C. stellatoidea) may occur.Antifungal susceptibility: Candida albicans (Australian national data); MIC µg/mL. Antifungal No ≤0.016 0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 32 ≥64 AMB 3013 2 16 162 650 679 1083 415 6 FLU 3016 2 2 3 132 1019 1223 411 85 26 19 35 15 44 ISAV 1036 955 59 8 5 2 3 1 1 1 1 VORI 2732 2290 227 86 31 29 31 16 10 2 4 6 POSA 2382 1039 978 228 66 31 23 14 1 2 ITRA 3016 178 814 1275 589 70 52 15 3 1 19 ANID 2111 678 773 479 172 6 1 1 1 MICA 2107 1976 109 14 5 2 1 CAS 1865 27 381 878 434 124 17 1 5FC 3016 4 147 1526 570 307 286 96 26 13 10 3 3 25 -
Candida auris
Candida auris is an emerging multidrug resistant yeast that causes invasive infections, that was first described in 2009 in Japan and has since been reported from numerous countries (Lockhart et al., 2017a,b; Jeffery-Smith et al., 2018; Spivak and Hanson, 2018; Heath et al., 2019; Zhu et al., 2020). Infections and outbreaks caused by C. auris in hospital settings have been rising over the past several years. Difficulty in its identification, multidrug resistance properties, associated high mortality rates, and long-term survival on surfaces in the environment make C. auris particularly problematic in clinical settings (Du et al., 2020). Candida auris is now a notifiable infection under public health regulations in many countries including Australia.

Candida auris grown on CHROMagar™ Candida Plus agar showing characteristic blue halo.
Candida auris may be misidentified as C. haemuloni or other yeast species using conventional phenotypic and biochemical methods (Kathuria et al., 2015; Jeffery-Smith et al., 2018). However, unlike these other Candida species, C. auris grows at 42oC, and this has become a useful differential characteristic (Casadevall et al., 2019). Candida auris is best identified by MALDI-ToF MS or by molecular methods (Du et al., 2020; Zhu et al., 2020).
MALDI-ToF MS: Can accurately differentiate Candida auris from other fungal species; however, accurate identification is dependent on the reference database being used (Kathuria et al., 2015; Jeffery-Smith et al., 2018; Zhu et al., 2020).
Molecular identification: ITS and D1/D2 sequencing provide accurate species identification (Kathuria et al., 2015; Kordalewska et al., 2017; Leach et al., 2018; Du et al., 2020; Zhu et al., 2020).
RG-2 organism.
Culture:
Colonies (SDA) white to cream-coloured smooth, glabrous, yeast-like.Microscopy:
Spherical to subspherical budding blastoconidia, 2-3 x 2-5 µm in size.India ink preparation:
Negative - no capsules present.Dalmau plate culture:
Ovoid budding yeast cells only. No pseudohyphae produced.Physiological tests: (+ Positive, - Negative, v Variable, w Weak, s Slow).
Fermentation:
Glucose + Sucrose + Lactose - Galactose - Maltose - Trehalose w.+ Growth reactions:
Glucose
+
l-Sorbose
-
myo-Inositol
-
Sucrose
+
l-Rhamnose
-
dl-Lactate
-
Raffinose
+
d-Xylose
-
d-Gluconate
-
Melibiose
-
l-Arabinose
-
2-Keto-d-gluconate
-
Galactose
-
d-Arabinose
-
d-Glucosamine
-
Lactose
-
d-Ribose
-
N-Acetyl-d-glucosamine
+
Trehalose
+
Glycerol
-
d-Glucuronate
-
Maltose
+
Erythritol
-
Nitrate
-
Melezitose
+
Ribitol
w,+
Urease
-
Methyl-⍺-d-glucoside
-
Galactitol
+
0.1% Cycloheximide
-
Soluble starch
+
d-Mannitol
+
Growth at 37oC
+
Cellobiose
-
d-Glucitol
+
Growth at 42oC
+,w,s
Key features:
Candida auris is difficult to identify using conventional phenotypic and biochemical methods and has often been misidentified as Candida haemuloni or Saccharomyces cerevisiae. Growth at 42oC on CHROMagar™ Candida produces white, pink, or dark purple colonies (Kumar et al., 2017), however on CHROMagar™ Candida Plus, C. auris colonies display a differential light blue colour with a blue halo (Mulet Bayona et al., 2022). Reliable identification methods are MALDI-ToF MS or ITS and D1/D2 sequencing.
Antifungal susceptibility: Candida auris (Chowdhary et al., 2018; and Australian national data); MIC µg/mL. Antifungal No ≤0.016 0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 32 ≥64 AMB 369 1 27 129 174 22 11 2 FLU 369 5 1 5 2 25 62 269 ISAV 360 44 138 53 38 37 26 11 8 5 VORI 369 21 23 94 91 43 36 28 19 4 10 POSA 369 97 90 85 48 31 8 3 3 2 2 ITRA 369 1 66 79 104 72 28 16 8 1 3 ANID 369 3 28 99 75 112 37 5 1 1 8 MICA 369 26 109 140 61 17 5 1 1 5 4 5FC 369 213 44 5 24 2 11 6 5 9 50 -
Candida dubliniensis
Candida dubliniensis is an occasional cause of candidaemia and mucosal infection, especially in HIV patients.
RG-2 organism.
Culture:
Colonies (SDA) white to cream-coloured smooth, glabrous, yeast-like.Microscopy:
Spherical to subspherical budding blastoconidia, 3-8 x 2-7 µm in size.India ink preparation:
Negative - no capsules present.Dalmau plate culture:
Branched pseudohyphae with dense verticils of blastoconidia and spherical, mostly terminal chlamydospores.Physiological Tests: + Positive, - Negative, v Variable, w Weak, s Slow, nd No Data Germ Tube + L-Sorbose - L-Arabinose - D-Glucitol + Fermentation Sucrose + D-Arabinose - 𝝰-M-D-glucoside +,s Glucose + Maltose + D-Ribose - D-Gluconate - Galactose +,s Cellobiose - L-Rhamnose - DL-Lactate + Sucrose - Trehalose s,+ D-Glucosamine v myo-Inositol - Maltose + Lactose - NAD-glucosamine + 2-K-D-Gluconate + Lactose - Melibiose - Glycerol w,s,+ D-Glucuronate - Trehalose v Raffinose - Erythritol - Nitrate - Assimilation Melezitose w,+ Ribitol + Urease - Glucose + Soluble Starch w,+ Galactitol - 0.1% Cycloheximide + Galactose + D-Xylose s,+ D-Mannitol + Growth at 40C + Key features:
Germ tube positive, similar to C. albicans, except for absence of growth at 42C; glycerol (mostly +), methyl-a-D-glucoside (-), trehalose (-), and D-xylose (-). Initial colonies dark green on CHROMagar and producing rough colonies on bird seed agar. ITS sequencing and MALDI-TOF can reliably distinguish C. dubliniensis from C. albicans.Antifungal susceptibility: Candida dubliniensis (Australian national data with additional ISAV data from Pfaller et al., 2013a); MIC µg/mL. Antifungal No ≤0.016 0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 32 ≥64 AMB 242 1 1 3 44 103 84 6 FLU 242 118 90 29 5 ISAV 75 70 4 1 VORI 239 239 POSA 231 125 77 26 3 ITRA 242 95 66 61 16 4 ANID 216 33 43 59 69 6 6 MICA 216 99 93 16 2 6 CAS 155 10 74 51 10 1 1 7 5FC 242 12 201 21 5 2 1 -
Candida haemuloni complex
Candida haemuloni has recently been reclassified as a complex of three phenotypically identical but genotypically distinct entities: C. haemuloni, C. duobushaemulonis and C. haemulonii var. vulneris, based on ITS and D1/D2 sequencing (Cendejas-Bueno et al. 2012, Ramos et al. 2015).
Note: Candida haemuloni and C. haemulonis are orthographic variants, but C. haemuloni is considered the correct name
Candida haemuloni
RG-1 organism.
Culture:
Colonies (SDA) white to cream-coloured smooth, glabrous, yeast-like.Microscopy:
Ovoid to globose, budding yeast-like cells or blastoconidia, 2-7 x 2-7 µm. No pseudohyphae produced.India ink preparation:
Negative - no capsules present.Dalmau plate culture:
No pseudohyphae produced.Physiological Tests: + Positive, - Negative, v Variable, w Weak, s Slow, nd No Data Germ Tube - L-Sorbose - L-Arabinose - D-Glucitol + Fermentation Sucrose + D-Arabinose -- 𝝰-M-D-Glucoside - Glucose + Maltose + D-Ribose - D-Gluconate + Galactose - Cellobiose - L-Rhamnose +,w DL-Lactate - Sucrose - Trehalose + D-Glucosamime +,s myo-Inositol - Maltose - Lactose - N-A-D-glucosamine + 2-K-D-Gluconate + Lactose - Melibiose - Glycerol +,s D-Glucuronate - Trehalose +,s Raffinose +,s Erythritol - Nitrate - Assimilation Melezitose +,w Ribitol +,s Urease - Glucose + Soluble Starch - Galactitol - 0.1% Cycloheximide - Galactose +,w D-Xylose - D-Mannitol + Growth at 37C - Key features: Germ tube negative yeast and sugar assimilation pattern. Molecular identification may be required. C. haemulonii has been reported from a few cases of fungaemia but clinical isolations remain rare.
Antifungal susceptibility: Candida haemulonii complex (Australian national data); MIC µg/mL. Antifungal No ≤0.016 0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 32 ≥64 AMB 30 2 10 7 7 1 2 FLU 30 1 3 1 25 VORI 28 1 1 1 1 20 4 POSA 23 1 2 1 1 3 16 ITRA 30 2 1 2 1 24 ANID 21 13 1 3 1 1 2 MICA 21 2 10 3 2 4 CAS 28 1 2 4 6 1 14 5FC 25 2 4 7 6 2 3 1 -
Candida inconspicua
Candida inconspicua is a rare cause of candidaemia.
RG-2 organism.
Culture:
Colonies (SDA) white to cream-coloured smooth, glabrous, yeast-like.Microscopy:
Ellipsoidal budding blastoconidia, 3-5 x 1.8-3 µm in size. No pseudohyphae or chlamydospores produced.India ink preparation:
Negative - no capsules present.Dalmau plate culture:
No pseudohyphae produced.Physiological Tests: + Positive, - Negative, v Variable, w Weak, s Slow, nd No Data Germ Tube - L-Sorbose - L-Arabinose - D-Glucitol - Fermentation Sucrose D-Arabinose - 𝝰-M-D-Glucoside - Glucose + Maltose - D-Ribose - D-Gluconate - Galactose - Cellobiose - L-Rhamnose - DL-Lactate + Sucrose - Trehalose - D-Glucosamime + myo-Inositol - Maltose - Lactose - N-A-D-glucosamine + 2-K-D-Gluconate - Lactose - Melibiose - Glycerol + D-Glucuronate - Trehalose - Raffinose - Erythritol - Nitrate - Assimilation Melezitose - Ribitol - Urease - Glucose + Soluble Starch - Galactitol - 0.1% Cycloheximide - Galactose - D-Xylose - D-Mannitol - Growth at 40C + Key features:
Germ tube negative yeast and sugar assimilation pattern and colonies are white on Candida CHROMagar.Antifungal susceptibility: Candida inconspicua (Australian national data); MIC µg/mL. Antifungal No ≤0.016 0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 32 ≥64 AMB 17 4 1 1 4 6 1 FLU 17 2 5 10 VORI 14 2 4 3 3 1 POSA 14 2 3 5 3 1 ITRA 16 2 6 7 1 ANID 4 3 1 MICA 4 3 1 5FC 15 1 1 2 5 3 1 1 1 -
Candida parapsilosis complex
Recently Candida parapsilosis has been recognised as a complex of four species: C. parapsilosis, C. orthopsilosis, C. metapsilosis and Lodderomyces elongisporus (Tavanti et al. 2005).
These four species are phenotypically indistinguishable and are best identified by ITS sequencing or MALDI-TOF MS analysis.
Candida parapsilosis Candida metapsilosis Candida orthopsilosis
-
Candida tropicalis
Candida tropicalis is a major cause of septicaemia and disseminated candidiasis. It is also found as part of the normal human mucocutaneous flora and environmental isolations have been made from faeces, shrimp, kefir and soil.
RG-2 organism.
Culture:
Colonies (SDA) white to cream-coloured smooth, glabrous, yeast-like.Microscopy:
Spherical to subspherical budding yeast-like cells or blastoconidia, 3.5-7 x 5.5-10 µm.India ink preparation:
Negative - no capsules present.Dalmau plate culture:
Abundant, long, wavy, branched pseudohyphae with numerous ovoid blastoconidia, budding off. Terminal vesicles (chlamydospores) are not produced.Physiological Tests: + Positive, - Negative, v Variable, w Weak, s Slow, nd No Data Germ Tube - L-Sorbose v L-Arabinose - D-Glucitol + Fermentation Sucrose v D-Arabinose - 𝝰-M-D-Glucoside v Glucose + Maltose + D-Ribose v,s D-Gluconate v Galactose + Cellobiose v L-Rhamnose - DL-Lactate v Sucrose v Trehalose + D-Glucosamime v myo-Inositol - Maltose + Lactose - N-A-D-glucosamine + 2-K-D-Gluconate + Lactose - Melibiose - Glycerol v D-Glucuronate - Trehalose +,s Raffinose - Erythritol - Nitrate - Assimilation Melezitose v Ribitol v Urease - Glucose + Soluble Starch + Galactitol - 0.1% Cycloheximide + Galactose + D-Xylose + D-Mannitol + Growth at 40C + Key features:
Germ tube negative yeast and sugar assimilation pattern. Colonies are dark blue on Candida CHROMagar.Antifungal susceptibility: Candida tropicalis (Australian national data with additional ISAV data from Pfaller et al., 2013a); MIC µg/mL. Antifungal No ≤0.016 0.03 0.06 0.125 0.25 0.5 1 2 4 8 16 32 ≥64 AMB 441 5 41 77 129 165 23 1 FLU 441 1 9 59 161 123 46 15 6 5 17 ISAV 218 13 51 64 48 20 14 5 1 1 1 VORI 409 22 47 104 126 66 18 7 9 2 7 1 POSA 390 8 32 66 132 107 27 10 2 6 ITRA 442 2 12 23 163 176 50 4 2 1 1 8 ANID 336 17 22 60 204 29 2 1 1 MICA 336 62 230 36 3 1 2 2 5FC 442 31 283 81 24 5 2 1 1 2 1 2 9
Quick Links to previous yeasts described as Candida species
Nakaseomyces glabratus (formerly C. glabrata)
Pichia kudriavzevii (formerly Candida krusei)
Meyerozyma guilliermondii (formerly Candida guilliermondii)
Clavispora lusitaniae (formerly Candida lusitaniae)
Kluyveromyces marxianus (formerly Candida kefyr)
Diutina catenulata (formerly Candida catenulata)
Diutina rugosa (formerly Candida rugosa)
Wickerhamomyces anomalus (formerly Candida pelliculosa)
